```{r include=FALSE}
require(Hmisc)
options(qproject='rms', prType='html')
require(qreport)
getRs('qbookfun.r')
hookaddcap()
knitr::set_alias(w = 'fig.width', h = 'fig.height', cap = 'fig.cap', scap ='fig.scap')
```
# Transform-Both-Sides Regression {#sec-areg}
## Background
* Challenges of traditional modeling (e.g. OLS) for estimation or inference
+ How should continuous predictors be transformed so as to get a
good fit?
+ Is it better to transform the response variable? How does one
find a good transformation that simplifies the right-hand side
of the equation?
+ What if $Y$ needs to be transformed non-monotonically (e.g.,
$|Y - 100|$) before it will have any
correlation with $X$?
* Challenges from need for normality and equal variance assumptions to draw accurate inferences
+ If for the untransformed original scale of the response $Y$
the distribution of the residuals is not normal with constant
spread, ordinary methods will not yield correct inferences
(e.g., confidence intervals will not have the desired coverage
probability and the intervals will need to be asymmetric).
+ Quite often there is a transformation of $Y$ that will yield
well-behaving residuals. How do you find this
transformation? Can you find a transformation for the $X$s at
the same time?
+ All classical statistical inferential methods assume that the
full model was pre-specified, that is, the model was not
modified after examining the data. How does one correct
confidence limits, for example, for data-based model and
transformation selection?
## Generalized Additive Models
* @has90gen developed GAMs for a variety of $Y$ distributions
* Most GAMs are highly parametric, just relaxing assumptions about $X$
* Nonparametrically estimate all $X$ transformations
* GAMs assume $Y$ is already well transformed
* Excellent software for GAMs: `R` packages `gam`, `mgcv`, `robustgam`.
## Nonparametric Estimation of $Y$-Transformation
There are a few approaches that also estimate the $Y$-transformation
### ACE
* @bre85est: _alternating conditional expectation_ (ACE)
* Simultaneously transforms $X$ and $Y$ to maximize $R^2$
$$g(Y) = f_{1}(X_{1}) + f_{2}(X_{2}) + \ldots + f_{p}(X_{p})$$ {#eq-areg-aceavas}
* Allows analyst to impose monotonicity restrictions on transformations
* Categorical $X$ automatically numerically scored
* $Y$-transform allowed to be non-monotonic
+ can create an identifiability problem
* Estimates _maximal correlation_ between $X$ and $Y$
* Basis for nonparametric estimation for continuous $X$: "super smoother" (R `supsmu` function)
### AVAS
* @tib88est: _additivity and variance stabilization_ (AVAS)
* Forces $g(Y)$ to be monotonic
* Maximizes $R^2$ while forcing $g(Y)$ to have nearly constant variance of residuals
* Model specification still @eq-areg-aceavas
### Overfitting
* Estimating so many transformations, especially $g(Y)$ effectively adds many parameters to the model
* Results in overfitting ($R^2$ inflation)
* Also no inferential measures provided
* Use the bootstrap to correct apparent $R^2$ ($R^{2}_{app}$) for overfitting
* Estimates the optimism (bias) in $R^{2}_{app}$ and subtracts this optimism from
$R^{2}_{app}$ to get an optimism-corrected estimate
* Also used to compute confidence limits for all estimated transformations
* And for predictor partial effects
* Must repeat **all** supervised learning steps afresh for each resample
* Limited testing has shown that the sample size needs to exceed 100 for
ACE and AVAS to provide stable estimates
## Obtaining Estimates on the Original Scale
* Traditional approach: take transformations of $Y$ before fitting
* Logarithm is the most common transformation.^[A disadvantage of transform-both-sides regression is this difficulty of interpreting estimates on the original scale. Sometimes the use of a special generalized linear model can allow for a good fit without transforming $Y$.]
* If residuals on $g(Y)$ have median 0, inverse transformed predicted values estimate median $Y | X$ (quantiles are transformation-preserving, unlike mean)
* How to estimate general parameters such as means on original $Y$ scale?
* Easy of residuals known to be Gaussian
* More general: @dua83sme "smearing estimator"
* One-sample case, log transformation
+ $\hat{\theta} = \sum_{i=1}^{n} \log(Y_{i})/n$,
+ residuals from this fitted value are given by $e_{i} = \log(Y_{i}) - \hat{\theta}$
+ smearing estimator of the population mean is $\sum \exp(\hat{\theta} + e_{i})/n$
+ is the ordinary sample mean $\overline{Y}$
* Smearing estimator needed when doing regression
* Regression run on $g(Y)$
+ estimated values $\hat{g}(Y_{i}) = X_{i}\hat{\beta}$
+ residuals on transformed scale $e_{i} = \hat{g}(Y_{i}) - X_{i}\hat{\beta}$
* Without restricting ourselves to estimating the population mean,
let $W(y_{1}, y_{2}, \ldots, y_{n})$ denote any function of a vector
of untransformed response values
* To estimate the population mean in the homogeneous one-sample case, $W$ is the simple average of all of
its arguments
* To estimate the population 0.25 quantile, $W$ is the
sample 0.25 quantile of $y_{1}, \ldots, y_{n}$
* Smearing estimator of the population parameter estimated by $W$ given $X$ is
$W(g^{-1}(a + e_{1}), g^{-1}(a + e_{2}), \ldots, g^{-1}(a +
e_{n}))$, where $g^{-1}$ is the inverse of the $g$ transformation and $a
= X\hat{\beta}$
* With AVAS algorithm, monotonic transformation $g$ is
estimated from the data
* Predicted value of $\hat{g}(Y)$ from @eq-areg-aceavas
* Extend the smearing estimator as $W(\hat{g}^{-1}(a + e_{1}), \ldots, \hat{g}^{-1}(a + e_{n}))$, where $a$ is the predicted transformed response given $X$
* $\hat{g}$ is nonparametric (i.e., a table look-up)
* R `areg.boot` function computes $\hat{g}^{-1}$ using reverse linear interpolation
* If assume residuals from $\hat{g}(Y)$ assumed to be symmetrically distributed,
their population median is zero
* Then can estimate the median on the untransformed scale by computing $\hat{g}^{-1}(X\hat{\beta})$ ^[To be safe, `areg.boot` adds the median residual to $X\hat{\beta}$ when estimating the population median (the median residual can be ignored by specifying `statistic='fitted'` to functions that operate on objects created by `areg.boot`)].
* For estimating quantiles of $Y$, quantile regression [@koe78reg] is more direct; see @aus05tut for a case study
## `R` Functions
* `R` `acepack` package: `ace` and `avas` functions
* `Hmisc` `areg.boot`: R modeling language notation and also implements a parametric spline version; estimates partial effects on $g(Y)$ and $Y$ varying one of the $X$s over two values
* Resamples every part of modeling process, as with @far92cos
* `monotone` function restricts a variable's
transformation to be monotonic
* `I` function restricts it to be linear
```{r eval=FALSE}
f <- areg.boot(Y ~ monotone(age) +
sex + weight + I(blood.pressure))
plot(f) #show transformations, CLs
Function(f) #generate S functions
#defining transformations
predict(f) #get predictions, smearing estimates
summary(f) #compute CLs on effects of each X
smearingEst() #generalized smearing estimators
Mean(f) #derive S function to
#compute smearing mean Y
Quantile(f) #derive function to compute smearing quantile
```
The methods are best described in a case study.
## Case Study {#sec-areg-case}
* Simulated data where conditional distribution of $Y$ is
log-normal given $X$, but where transform-both-sides regression
methods use un-logged $Y$
* Predictor $X_1$ is linearly related to log $Y$
* $X_2$ is related by $|X_{2} - \frac{1}{2}|$
* Categorical $X_3$ has reference group $a$ effect of
zero, group $b$ effect of 0.3, and group $c$ effect of 0.5
```{r sim}
require(rms)
set.seed(7)
n <- 400
x1 <- runif(n)
x2 <- runif(n)
x3 <- factor(sample(c('a','b','c'), n, TRUE))
y <- exp(x1 + 2*abs(x2 - .5) + .3*(x3=='b') + .5*(x3=='c') +
.5*rnorm(n))
# For reference fit appropriate OLS model
print(ols(log(y) ~ x1 + rcs(x2, 5) + x3), coefs=FALSE)
```
* Use 300 bootstrap resamples and run `avas`
* Only first 20 bootstraps are plotted
* Had we restricted `x1` to be linear would have specified `I(x1)`
```{r aregboot,results='hide'}
f <- areg.boot(y ~ x1 + x2 + x3, method='avas', B=300)
```
```{r}
f
```
* Coefficients hard to interpret as scale of transformations arbitrary
* Model was very slightly overfitted ($R^2$ dropped from
`r round(f$rsquared.app,2)` to `r round(f$rsquared.val,2)`),
and $R^2$ are in agreement with the OLS model fit
* Plot the transformations, 0.95 confidence bands, and a sample
of the bootstrap estimates
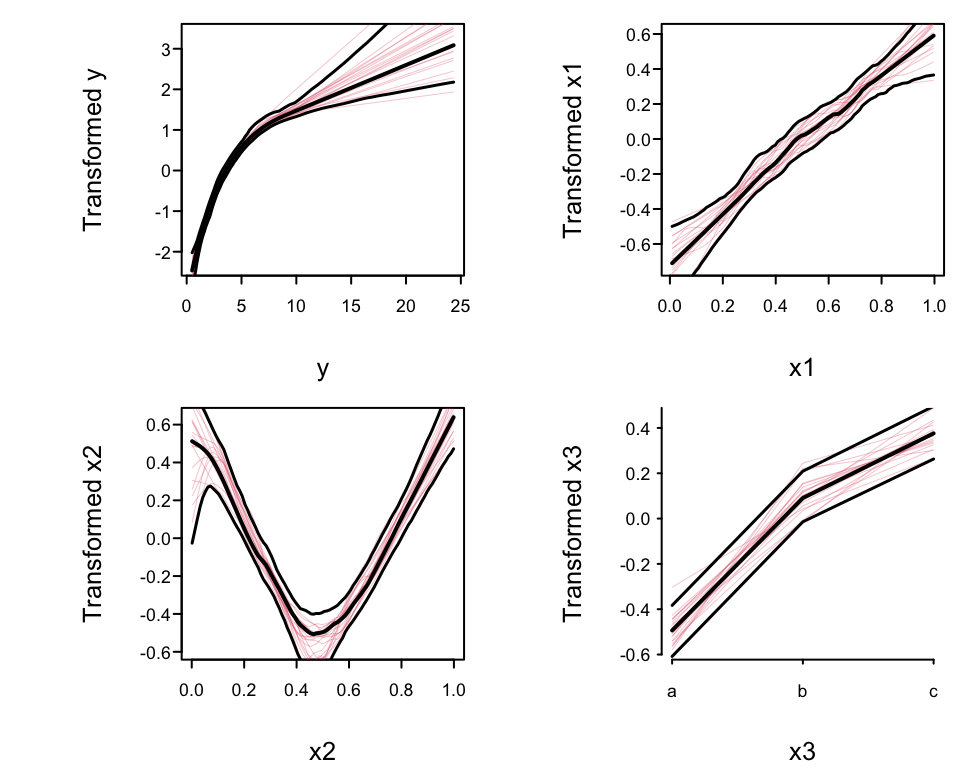
```{r trans,h=4,w=5,cap='`avas` transformations: overall estimates, pointwise $0.95$ confidence bands, and $20$ bootstrap estimates (red lines).',scap='Transformations estimated by `avas`'}
#| label: fig-areg-trans
spar(mfrow=c(2,2), ps=10)
plot(f, boot=20)
```
* Nonparametrically estimated transformation of `x1` almost linear
* Transformation of `x2` is close to $|x2 - 0.5|$
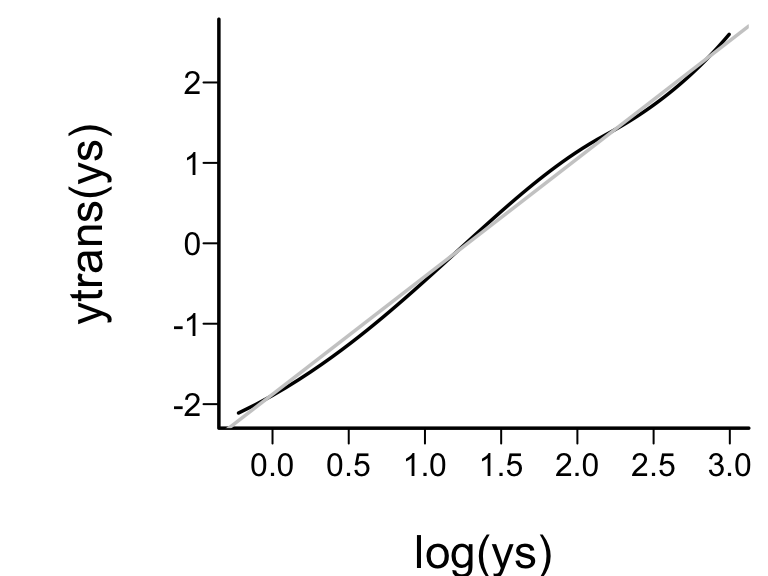
* True transformation of `y` is $\log(y)$, so variance
stabilization and normality of residuals will be achieved if the
estimated `y`-transformation is close to $\log(y)$.
```{r ytrans,w=4,h=3,cap='Checking estimated against optimal transformation'}
#| label: fig-areg-ytrans
spar(bty='l')
ys <- seq(.8, 20, length=200)
ytrans <- Function(f)$y # Function outputs all transforms
plot(log(ys), ytrans(ys), type='l')
abline(lm(ytrans(ys) ~ log(ys)), col=gray(.8))
```
* Approximate linearity = estimated transformation $\log$-like.^[Beware that use of a data--derived transformation in an ordinary model, as this will result in standard errors that are too small. This is because model selection is not taken into account. [@far92cos]]
* Obtain approximate tests of effects of each predictor
* `summary` sets all other predictors to reference values (e.g., medians)
* Compares predicted responses for a given level of the predictor $X$ with predictions for lowest setting of $X$
* Default predicted response for `summary` is the median; tests are for differences in medians
```{r }
summary(f, values=list(x1=c(.2, .8), x2=c(.1, .5)))
```
* Example: when `x1` increases from 0.2 to 0.8 predict an
increase in median `y` by 1.55 with bootstrap standard error 0.21, when all
other predictors are held to constants^[Setting them to other constants will yield different estimates of the `x1` effect, as the transformation of `y` is nonlinear.]
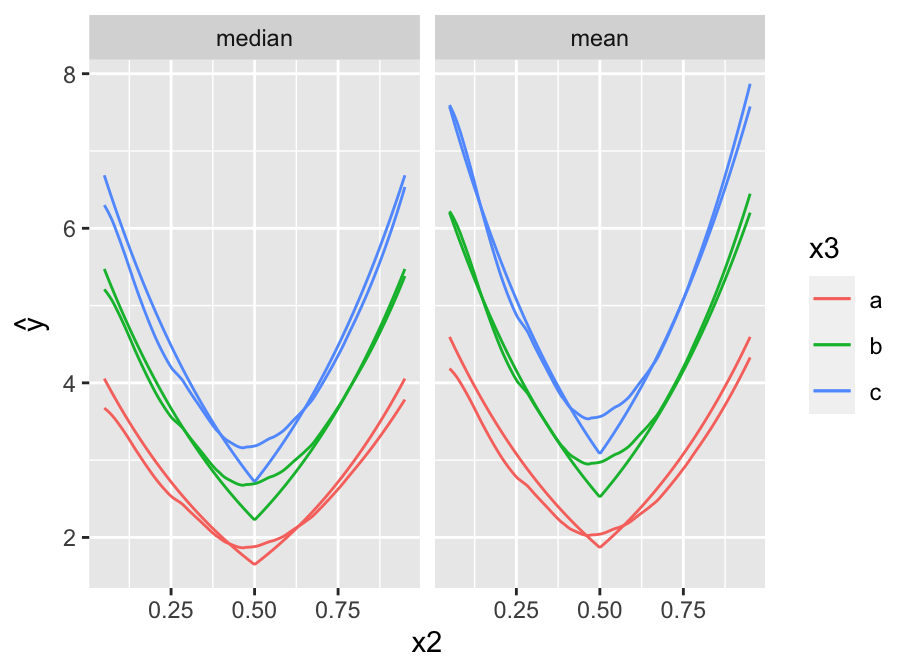
* Depict the fitted model by plotting predicted values
* `x2` varies on $x$-axis, 3 curves correspond to three values of `x3`
* `x1` set to 0.5
* Show estimates of both the median and the mean `y`.
```{r pred,h=3.5,w=4.75,cap='Predicted median (left panel) and mean (right panel) `y` as a function of `x2` and `x3`. True population curves are pointed.',scap='Predicted `y` as a function of `x2` and `x3`'}
#| label: fig-areg-pred
require(ggplot2)
newdat <- expand.grid(x2=seq(.05, .95, length=200),
x3=c('a','b','c'), x1=.5,
statistic=c('median', 'mean'))
yhat <- c(predict(f, subset(newdat, statistic=='median'),
statistic='median'),
predict(f, subset(newdat, statistic=='mean'),
statistic='mean'))
newdat <-
upData(newdat,
lp = x1 + 2*abs(x2 - .5) + .3*(x3=='b') +
.5*(x3=='c'),
ytrue = ifelse(statistic=='median', exp(lp),
exp(lp + 0.5*(0.5^2))), print=FALSE)
ggplot(newdat, aes(x=x2, y=yhat, col=x3)) + geom_line() +
geom_line(aes(x=x2, y=ytrue, col=x3)) +
facet_wrap(~ statistic) + ylab(expression(hat(y)))
```
```{r echo=FALSE}
saveCap('16')
```