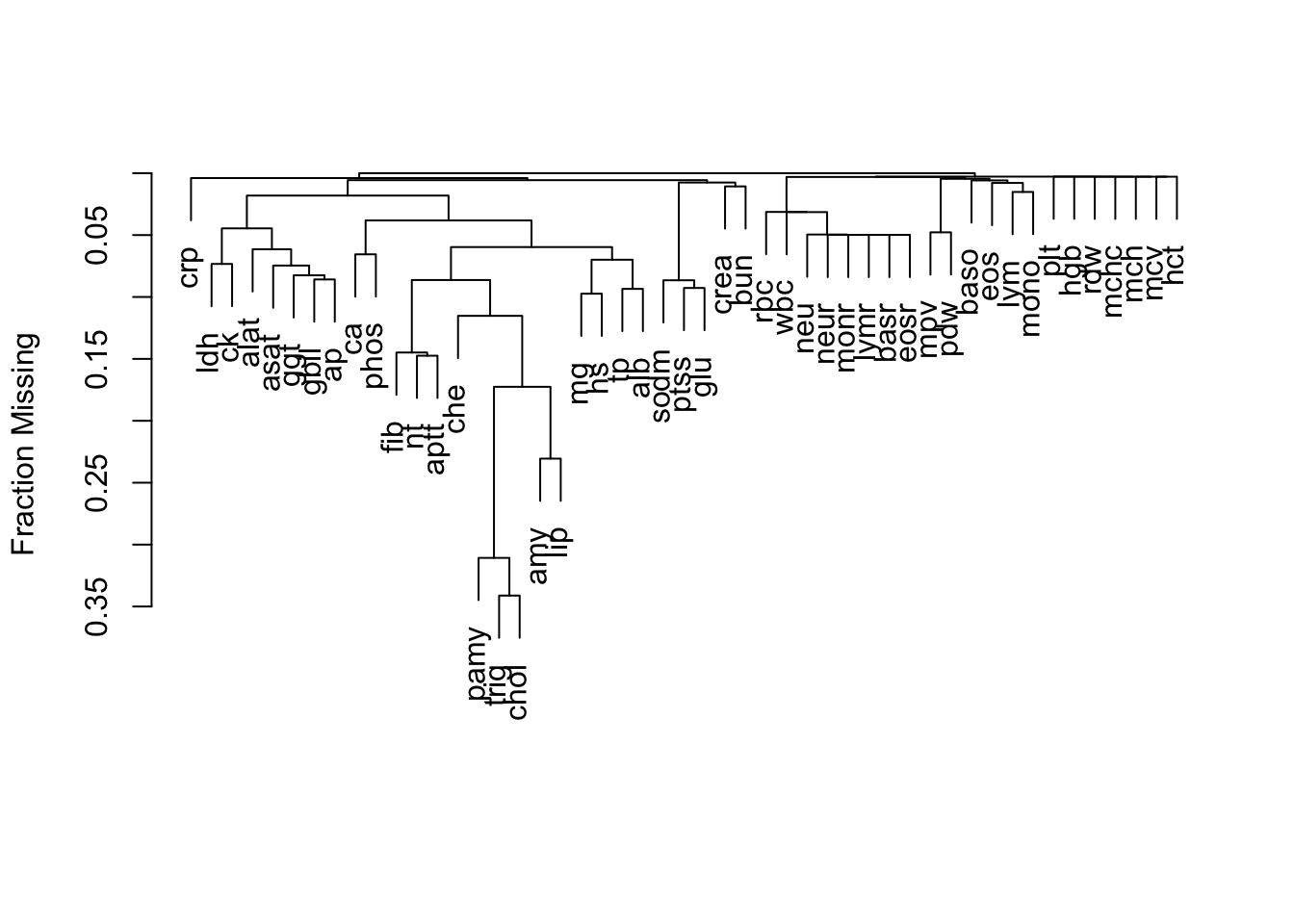
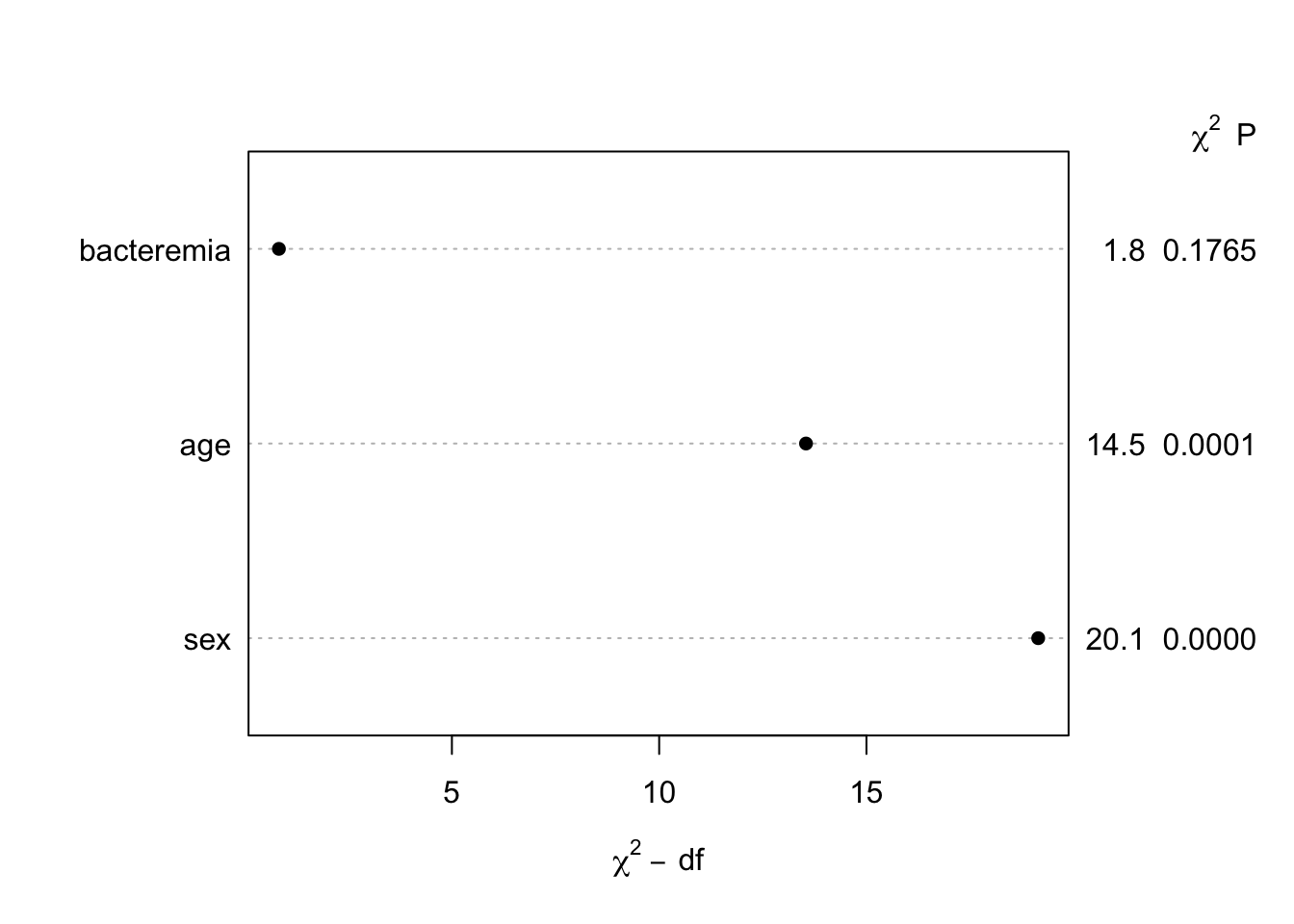
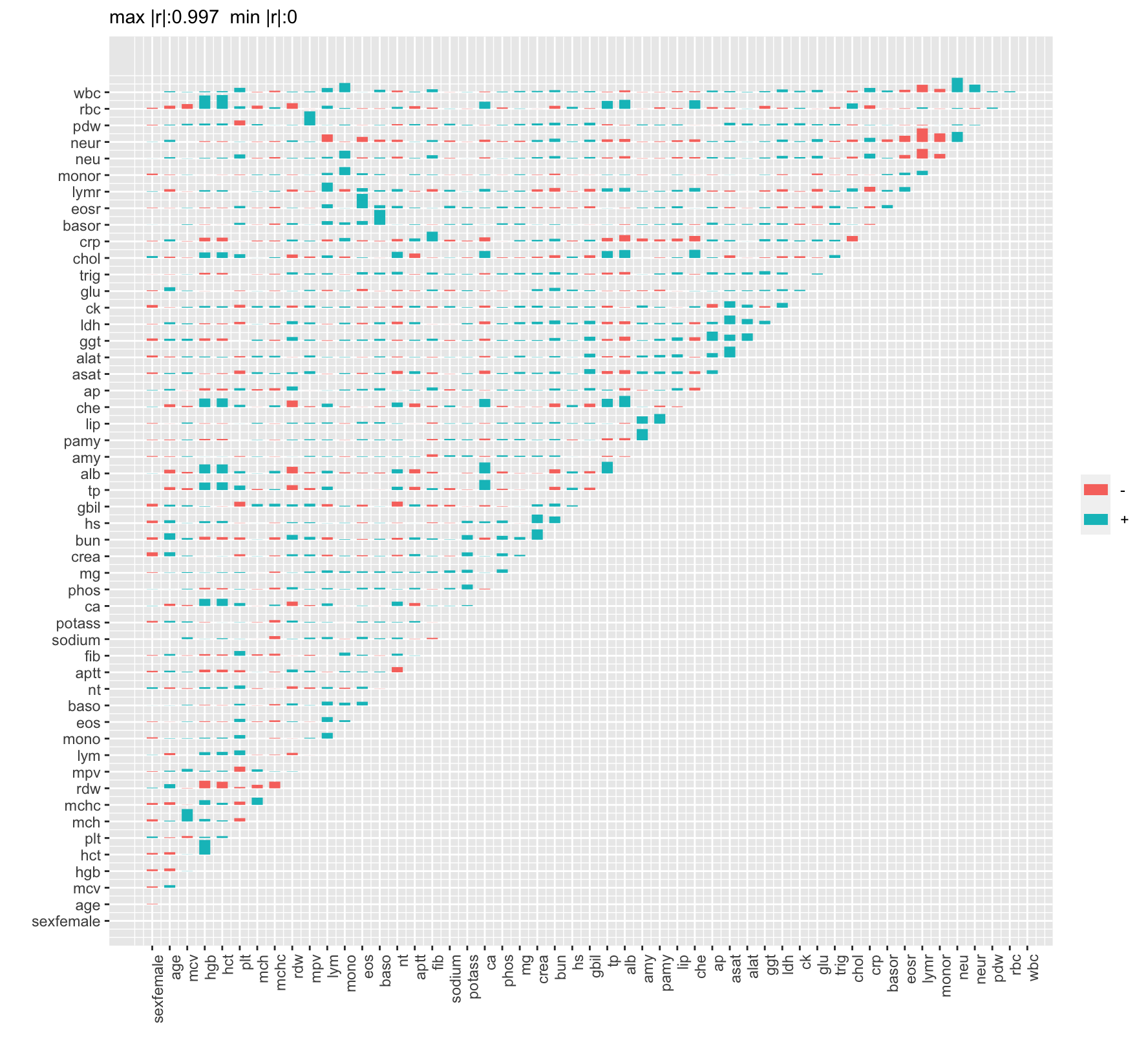
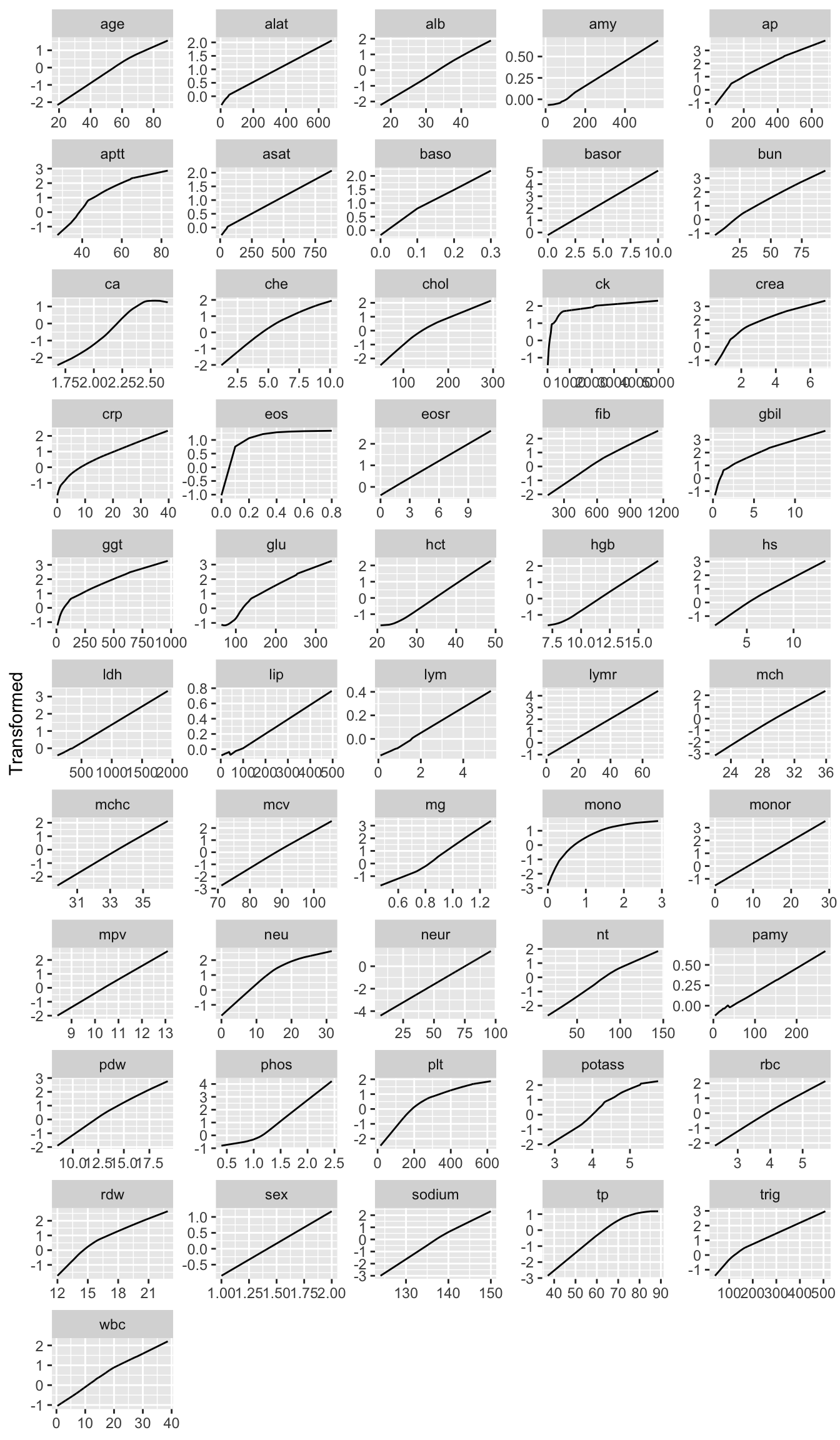
| id |
Patient Identification |
|
14691 |
0 |
14691 |
1.000 |
29353 |
29314 |
20965 |
|
2206 4834 13586 28755 44670 55598 59070 |
| age |
Patient Age |
years |
14691 |
0 |
85 |
1.000 |
56.17 |
56.5 |
20.78 |
|
24 29 43 58 70 79 84 |
| mcv |
Mean corpuscular volume |
pg |
14649 |
42 |
506 |
1.000 |
88.35 |
88.3 |
6.992 |
|
78.2 81.1 84.7 88.3 92.0 95.9 99.0 |
| hgb |
Haemoglobin |
G/L |
14650 |
41 |
157 |
1.000 |
11.57 |
11.5 |
2.558 |
|
8.2 8.8 9.9 11.4 13.2 14.6 15.4 |
| hct |
Haematocrit |
% |
14649 |
42 |
404 |
1.000 |
34.48 |
34.45 |
7.316 |
|
24.6 26.4 29.8 34.3 39.1 42.9 44.8 |
| plt |
Blood platelets |
G/L |
14649 |
42 |
718 |
1.000 |
220 |
209.5 |
130.1 |
|
50 81 140 204 277 369 445 |
| mch |
Mean corpuscular hemoglobin |
fl |
14649 |
42 |
232 |
1.000 |
29.58 |
29.65 |
2.693 |
|
25.3 26.7 28.4 29.7 31.0 32.4 33.4 |
| mchc |
Mean corpuscular hemoglobin concentration |
g/dl |
14649 |
42 |
124 |
0.999 |
33.47 |
33.5 |
1.546 |
|
31.1 31.7 32.6 33.5 34.4 35.2 35.6 |
| rdw |
Red blood cell distribution width |
% |
14635 |
56 |
173 |
1.000 |
15 |
14.7 |
2.385 |
|
12.4 12.7 13.4 14.5 16.0 18.0 19.5 |
| mpv |
Mean platelet volume |
fl |
13989 |
702 |
71 |
0.999 |
10.38 |
10.35 |
1.132 |
|
8.9 9.2 9.7 10.3 11.0 11.7 12.2 |
| lym |
Lymphocytes |
G/L |
14429 |
262 |
114 |
0.998 |
1.366 |
1.1 |
1.162 |
|
0.2 0.4 0.7 1.0 1.6 2.1 2.6 |
| mono |
Monocytes |
G/L |
14445 |
246 |
67 |
0.996 |
0.8527 |
0.8 |
0.5965 |
|
0.1 0.3 0.5 0.8 1.1 1.5 1.8 |
| eos |
Eosinophils |
G/L |
14556 |
135 |
36 |
0.867 |
0.1148 |
0.1 |
0.1585 |
|
0.0 0.0 0.0 0.1 0.1 0.3 0.4 |
| baso |
Basophils |
G/L |
14545 |
146 |
18 |
0.337 |
0.01725 |
0 |
0.03111 |
|
0.0 0.0 0.0 0.0 0.0 0.1 0.1 |
| nt |
Normotest |
% |
12224 |
2467 |
149 |
1.000 |
83.22 |
83.5 |
30.56 |
|
35 48 67 83 101 118 128 |
| aptt |
Activated partial thromboplastin time |
sec |
12142 |
2549 |
631 |
1.000 |
40.06 |
38.4 |
9.533 |
|
30.1 31.4 34.1 37.7 42.7 49.9 56.6 |
| fib |
Fibrinogen |
mg/dl |
12124 |
2567 |
1084 |
1.000 |
547.4 |
537 |
231 |
|
247 301 397 529 674 816 892 |
| sodium |
Sodium |
mmol/L |
13409 |
1282 |
58 |
0.994 |
137.2 |
137.5 |
5.034 |
|
129 132 135 137 140 142 144 |
| potass |
Potassium |
mmol/L |
12683 |
2008 |
408 |
1.000 |
4.003 |
3.97 |
0.6004 |
|
3.20 3.39 3.66 3.95 4.29 4.67 4.92 |
| ca |
Calcium |
mmol/L |
13415 |
1276 |
185 |
1.000 |
2.214 |
2.22 |
0.2213 |
|
1.89 1.96 2.09 2.22 2.35 2.45 2.51 |
| phos |
Phosphate |
mmol/L |
13449 |
1242 |
306 |
1.000 |
1.048 |
1.01 |
0.3993 |
|
0.55 0.64 0.81 0.99 1.20 1.47 1.74 |
| mg |
Magnesium |
mmol/L |
12822 |
1869 |
146 |
0.999 |
0.8136 |
0.81 |
0.1609 |
|
0.59 0.64 0.72 0.81 0.89 0.98 1.06 |
| crea |
Creatinine |
mg/dl |
14532 |
159 |
674 |
1.000 |
1.329 |
1.07 |
0.8518 |
|
0.620 0.690 0.810 1.000 1.350 2.160 3.144 |
| bun |
Blood urea nitrogen |
mg/dl |
14519 |
172 |
947 |
1.000 |
22.66 |
18.9 |
16.92 |
|
7.1 8.6 11.6 16.6 26.9 44.8 60.8 |
| hs |
Uric acid |
mg/dl |
11630 |
3061 |
169 |
1.000 |
5.413 |
5.2 |
2.625 |
|
2.2 2.7 3.7 5.0 6.6 8.5 10.0 |
| gbil |
Bilirubin |
mg/dl |
13250 |
1441 |
885 |
1.000 |
1.406 |
0.875 |
1.477 |
|
0.33 0.39 0.53 0.77 1.23 2.34 3.96 |
| tp |
Total protein |
G/L |
13108 |
1583 |
649 |
1.000 |
64.9 |
65.2 |
12.97 |
|
45.20 49.47 56.90 65.70 73.30 78.80 82.00 |
| alb |
Albumin |
G/L |
13015 |
1676 |
401 |
1.000 |
33.42 |
33.5 |
8.513 |
|
21.3 23.6 27.9 33.6 39.1 43.2 45.2 |
| amy |
Amylase |
U/L |
10778 |
3913 |
488 |
1.000 |
90.83 |
54.5 |
100.5 |
|
18 23 33 49 76 125 187 |
| pamy |
Pancreas amylase |
U/L |
7577 |
7114 |
280 |
0.999 |
41.66 |
25 |
47.28 |
|
7 9 14 22 36 64 97 |
| lip |
Lipases |
U/L |
10992 |
3699 |
444 |
1.000 |
63.82 |
26.5 |
89.88 |
|
6 8 14 23 40 79 135 |
| che |
Cholinesterase |
kU/L |
12244 |
2447 |
997 |
1.000 |
4.79 |
4.705 |
2.378 |
|
1.70 2.17 3.15 4.60 6.22 7.65 8.49 |
| ap |
Alkaline phosphatase |
U/L |
13291 |
1400 |
672 |
1.000 |
118.8 |
92 |
91.51 |
|
42 49 63 84 123 206 302 |
| asat |
Aspartate transaminase |
U/L |
13537 |
1154 |
650 |
1.000 |
86.9 |
38 |
115.6 |
|
15 17 22 31 56 121 218 |
| alat |
Alanin transaminase |
U/L |
13704 |
987 |
578 |
1.000 |
67.66 |
31.5 |
90.07 |
|
9 11 16 26 48 101 175 |
| ggt |
Gamma-glutamyl transpeptidase |
G/L |
13429 |
1262 |
858 |
1.000 |
115.1 |
68 |
141.3 |
|
13.0 16.0 25.0 49.0 117.0 262.2 429.0 |
| ldh |
Lactate dehydrogenase |
U/L |
12977 |
1714 |
1137 |
1.000 |
331.2 |
257.5 |
240.9 |
|
136 152 187 239 332 508 724 |
| ck |
Creatine kinase |
U/L |
12611 |
2080 |
1506 |
1.000 |
385 |
106.5 |
615.4 |
|
18 25 42 80 184 577 1155 |
| glu |
Glucose |
mg/dl |
10499 |
4192 |
389 |
1.000 |
126.4 |
117.5 |
48.3 |
|
78 85 97 113 138 177 216 |
| trig |
Triclyceride |
mg/dl |
9630 |
5061 |
538 |
1.000 |
141.7 |
124 |
90.33 |
|
54 64 83 115 165 241 307 |
| chol |
Cholesterol |
mg/dl |
9646 |
5045 |
339 |
1.000 |
150.8 |
147.5 |
59.23 |
|
74 89 113 145 182 219 243 |
| crp |
C-reactive protein |
mg/dl |
14536 |
155 |
3328 |
1.000 |
10.92 |
9.965 |
10.39 |
|
0.29 0.77 2.87 8.57 16.45 24.49 29.61 |
| basor |
Basophil ratio |
% |
13959 |
732 |
419 |
0.322 |
0.145 |
0 |
0.2679 |
|
0.0000 0.0000 0.0000 0.0000 0.0000 0.5501 1.0526 |
| eosr |
Eosinophil ratio |
% |
13959 |
732 |
927 |
0.891 |
1.297 |
0.8772 |
1.825 |
|
0.0000 0.0000 0.0000 0.5882 1.7857 3.4900 5.0000 |
| lymr |
Lymphocyte ratio |
% (mg/dl) |
13959 |
732 |
3121 |
1.000 |
14.61 |
12.53 |
11.87 |
|
2.752 4.000 6.757 11.340 18.182 27.869 36.620 |
| monor |
Monocyte ratio |
% |
13959 |
732 |
2334 |
1.000 |
8.793 |
8.227 |
5.4 |
|
2.000 3.390 5.634 8.000 10.870 14.141 17.021 |
| neu |
Neutrophiles |
G/L |
13963 |
728 |
374 |
1.000 |
8.367 |
7.75 |
5.776 |
|
1.60 2.70 4.60 7.30 10.80 15.08 18.40 |
| neur |
Neutrophile ratio |
% |
13959 |
732 |
3850 |
1.000 |
75.15 |
77.18 |
15.6 |
|
47.42 57.88 69.23 78.33 85.32 90.13 92.63 |
| pdw |
Platelet distribution width |
% |
13589 |
1102 |
167 |
1.000 |
12.29 |
12.1 |
2.375 |
|
9.3 9.8 10.8 12.0 13.4 15.1 16.4 |
| rbc |
Red blood count |
T/L |
14230 |
461 |
65 |
0.999 |
3.936 |
3.95 |
0.8772 |
|
2.7 2.9 3.4 3.9 4.5 4.9 5.2 |
| wbc |
White blood count |
G/L |
14229 |
462 |
2710 |
1.000 |
11.23 |
10.05 |
7.602 |
|
2.66 4.26 6.63 9.60 13.53 18.22 22.27 |