Code
a <- aregImpute(~ age + day_dis + day_follow_up, data=d,
constraint=list(day_dis=expression(d$day_dis < r$day_follow_up)))Consider a study of the effectiveness of a cardiac rehabilitation program. Suppose a sample size of N=300 will result in an acceptable margin of error in estimating the overall treatment effect. The investigators allow for 20% dropout so multiplies N by 1/0.8 = 1.25. What serious mistake are they likely making?
1 “Although missing at random (MAR) is a non-testable assumption, it has been pointed out in the literature that we can get very close to MAR if we include enough variables in the imputation models” Harel & Zhou (2007).
See Carpenter & Smuk (2021), Schafer & Graham (2002), Donders et al. (2006), Harel & Zhou (2007), Allison (2001), White et al. (2011), van Buuren (2012) for an introduction to missing data and imputation concepts. See Hazewinkel et al. (2023) for a way to indirectly detect MNAR by comparing variances of response variables between treatments.
2 Twisk et al. (2013) found instability in using multiple imputation of longitudinal data, and advantages of using instead full likelihood models.
3 White & Royston (2009) provide a method for multiply imputing missing covariate values using censored survival time data.
4 \(Y\) is so valuable that if one is only missing a \(Y\) value, imputation is not worthwhile, and imputation of \(Y\) is not advised if MCAR or MAR.
Deletion of records—
5 Multiple imputation of \(Y\) in that case does not improve the analysis and assumes the imputation model is correct.
Adding extra categories of categorical predictors—
6 E.g. you have a measure of marital happiness, dichotomized as high or low, but your sample contains some unmarried people. OK to have a 3-category variable with values high, low, and unmarried—Paul Allison, IMPUTE list, 4Jul09.
Likewise, serious problems are caused by setting missing continuous predictors to a constant (e.g., zero) and adding an indicator variable to try to estimate the effect of missing values.
Two examples from Donders et al. (2006) using binary logistic regression, \(N=500\).
Results of 1000 Simulations With \(\beta_{1}=1.0\) with MAR and Two Types of Imputation
| Imputation | \(\hat{\beta}_{1}\) | S.E. | Coverage of |
|---|---|---|---|
| Method | 0.90 C.I. | ||
| Single | 0.989 | 0.09 | 0.64 |
| Multiple | 0.989 | 0.14 | 0.90 |
Now consider a simulation with \(\beta_{1}=1, \beta_{2}=0\), \(X_{2}\) correlated with \(X_{1} (r=0.75)\) but redundant in predicting \(Y\), use missingness indicator when \(X_{1}\) is MCAR in 0.4 of 500 subjects. This is also compared with grand mean fill-in imputation.
Results of 1000 Simulations Adding a Third Predictor Indicating Missing for \(X_{1}\)}
| Imputation | \(\hat{\beta}_{1}\) | \(\hat{\beta}_{2}\) |
|---|---|---|
| Method | ||
| Indicator | 0.55 | 0.51 |
| Overall mean | 0.55 |
In the incomplete observations the constant \(X_{1}\) is uncorrelated with \(X_{2}\).
The goal of imputation is to preserve the information and meaning of the non-missing data.
There is a full Bayesian modeling alternative to all the methods presented below. The Bayesian approach requires more effort but has several advantages (Erler et al., 2016).
Exactly how are missing values estimated?
transcan function in Hmisc library: “optimal” transformations of all variables to make residuals more stable and to allow non-monotonic transformationsaregImpute function in Hmisc: good approximation to full Bayesian multiple imputation procedure using the bootstraptranscan and aregImpute use the following for fitting imputation models: NAs to median (mode for categoricals)aregImpute and transcan work with fit.mult.impute to make final analysis of response variable relatively easyNA. Key considerations are how toNAHmisc 5.1-1d$) and the recipient (target) observation (variables prefixed by r$) having the missing valuetranscan does not do that; aregImpute doesaregImpute algorithm (Moons et al., 2006)
R MICE approachSee Barzi & Woodward (2004) for a nice review of multiple imputation with detailed comparison of results (point estimates and confidence limits for the effect of the sometimes-missing predictor) for various imputation methods. Barnes et al. (2006) have a good overview of imputation methods and a comparison of bias and confidence interval coverage for the methods when applied to longitudinal data with a small number of subjects. Horton & Kleinman (2007) have a good review of several software packages for dealing with missing data, and a comparison of them with aregImpute. Harel & Zhou (2007) provide a nice overview of multiple imputation and discuss some of the available software. White & Carlin (2010) studied bias of multiple imputation vs. complete-case analysis. White et al. (2011) provide much practical guidance.
Caution: Methods can generate imputations having very reasonable distributions but still not having the property that final response model regression coefficients have nominal confidence interval coverage. It is worth checking that imputations generate the correct collinearities among covariates.
MICE and aregImpute we are using the chained equation approach (White et al., 2011) Hmisc fit.mult.impute function run with lrt=TRUE runs and saves all the needed LR tests to allow rms processMI to compute everything elseprocessMI computes \(\hat{f}\), the \(F\) test denominator d.f., and the \(\chi^2\) discount factor and uses these to get the final imputation-adjusted LR testsfit.mult.impute computes very approximate Wald statistics by multiplying the variance-covariance matrix computed on the stacked data by \(M\) if individual completed dataset analyses are not donefit.mult.impute computes the final \(\hat{\beta}\) this way if stacking is done; however the variance-covariance matrix is estimated from Rubin’s rule as in the Wald approach first used in this chapterrms anova function has very litle overhead though, due to it only computing the design matrix once, and calling low-level fitting functions for all LRTs once the overall model is fitted with a high-level function.Example
require(rms)
set.seed(1)
n <- 500
x1 <- runif(n)
x2 <- runif(n)
L <- x1 + 2*x2
y <- rbinom(n, 1, plogis(L))
x2[1:300] <- NA # make 300 observations have missing x2
d <- data.frame(x1, x2, y)
a <- aregImpute(~ y + x1 + x2, n.impute=10, data=d, pr=FALSE)
f <- fit.mult.impute(y ~ x1 + x2, lrm, a, data=d, pr=FALSE)
fLogistic Regression Model
fit.mult.impute(formula = y ~ x1 + x2, fitter = lrm, xtrans = a,
data = d, pr = FALSE)
| Model Likelihood Ratio Test |
Discrimination Indexes |
Rank Discrim. Indexes |
|
|---|---|---|---|
| Obs 500 | LR χ2 48.78 | R2 0.150 | C 0.725 |
| 0 93 | d.f. 2 | R22,500 0.089 | Dxy 0.451 |
| 1 407 | Pr(>χ2) <0.0001 | R22,227.1 0.183 | γ 0.451 |
| max |∂log L/∂β| 2×10-5 | Brier 0.138 | τa 0.137 |
| β | S.E. | Wald Z | Pr(>|Z|) | |
|---|---|---|---|---|
| Intercept | -0.2395 | 0.4788 | -0.50 | 0.6169 |
| x1 | 1.6851 | 0.4804 | 3.51 | 0.0005 |
| x2 | 2.2036 | 0.8701 | 2.53 | 0.0113 |
In the above results, the LR \(\chi^2\) did not account for imputation and is too high. So are the \(R^2\) measures derived from it.
Compute Wald \(\chi^2\) test statistics accounting for imputation
Wald Statistics for y |
|||
| χ2 | d.f. | P | |
|---|---|---|---|
| x1 | 12.30 | 1 | 0.0005 |
| x2 | 6.41 | 1 | 0.0113 |
| TOTAL | 15.96 | 2 | 0.0003 |
Now compute LR test statistics accounting for imputation
Logistic Regression Model
fit.mult.impute(formula = y ~ x1 + x2, fitter = lrm, xtrans = a,
data = d, lrt = TRUE, pr = FALSE)
| Model Likelihood Ratio Test |
Discrimination Indexes |
Rank Discrim. Indexes |
|
|---|---|---|---|
| Obs 500 | LR χ2 45.74 | R2 0.142 | C 0.724 |
| 0 93 | d.f. 2 | R22,5000 0.087 | Dxy 0.449 |
| 1 407 | Pr(>χ2) <0.0001 | R22,2271.1 0.182 | γ 0.449 |
| max |∂log L/∂β| 4×10-6 | Brier 0.139 | τa 0.136 |
| β | S.E. | Wald Z | Pr(>|Z|) | |
|---|---|---|---|---|
| Intercept | -0.2047 | 0.4788 | -0.43 | 0.6690 |
| x1 | 1.6607 | 0.4804 | 3.46 | 0.0005 |
| x2 | 2.1519 | 0.8701 | 2.47 | 0.0134 |
Likelihood Ratio Statistics for y |
|||
| χ2 | d.f. | P | |
|---|---|---|---|
| x1 | 10.23 | 1 | 0.0014 |
| x2 | 6.30 | 1 | 0.0121 |
| TOTAL | 16.01 | 2 | 0.0003 |
| Imputation penalties | |||
| Test | Missing Information Fraction |
Denominator d.f. |
χ2 Discount |
|---|---|---|---|
| x1 | 0.336 | 79.5 | 0.664 |
| x2 | 0.788 | 14.5 | 0.212 |
| TOTAL | 0.650 | 42.6 | 0.350 |
The Chan & Meng \(F\) approximation (processMI uses the \(\chi^2\) version of it) is needed for x2 as the denominator d.f. of 14.5 is too low for \(p\)-values from the \(\chi^2\) distribution to well approximate those from the \(F\) distribution. Increase the number of imputations to increase the denominator d.f.
Logistic Regression Model
fit.mult.impute(formula = y ~ x1 + x2, fitter = lrm, xtrans = a,
data = d, lrt = TRUE, pr = FALSE)
| Model Likelihood Ratio Test |
Discrimination Indexes |
Rank Discrim. Indexes |
|
|---|---|---|---|
| Obs 500 | LR χ2 40.49 | R2 0.126 | C 0.713 |
| 0 93 | d.f. 2 | R22,25000 0.078 | Dxy 0.425 |
| 1 407 | Pr(>χ2) <0.0001 | R22,11355.3 0.163 | γ 0.425 |
| max |∂log L/∂β| 7×10-6 | Brier 0.141 | τa 0.129 |
| β | S.E. | Wald Z | Pr(>|Z|) | |
|---|---|---|---|---|
| Intercept | -0.0783 | 0.3986 | -0.20 | 0.8443 |
| x1 | 1.5982 | 0.4640 | 3.44 | 0.0006 |
| x2 | 1.9308 | 0.7341 | 2.63 | 0.0085 |
Likelihood Ratio Statistics for y |
|||
| χ2 | d.f. | P | |
|---|---|---|---|
| x1 | 12.10 | 1 | 0.0005 |
| x2 | 6.74 | 1 | 0.0094 |
| TOTAL | 17.51 | 2 | 0.0002 |
| Imputation penalties | |||
| Test | Missing Information Fraction |
Denominator d.f. |
χ2 Discount |
|---|---|---|---|
| x1 | 0.153 | 2097.1 | 0.847 |
| x2 | 0.724 | 93.4 | 0.276 |
| TOTAL | 0.568 | 304.2 | 0.432 |
What happens if we use an extremely high number of imputations?
Logistic Regression Model
fit.mult.impute(formula = y ~ x1 + x2, fitter = lrm, xtrans = a,
data = d, lrt = TRUE, pr = FALSE)
| Model Likelihood Ratio Test |
Discrimination Indexes |
Rank Discrim. Indexes |
|
|---|---|---|---|
| Obs 500 | LR χ2 42.41 | R2 0.132 | C 0.717 |
| 0 93 | d.f. 2 | R22,250000 0.081 | Dxy 0.433 |
| 1 407 | Pr(>χ2) <0.0001 | R22,113553 0.170 | γ 0.433 |
| max |∂log L/∂β| 0.0001 | Brier 0.140 | τa 0.131 |
| β | S.E. | Wald Z | Pr(>|Z|) | |
|---|---|---|---|---|
| Intercept | -0.1123 | 0.3900 | -0.29 | 0.7734 |
| x1 | 1.5950 | 0.4688 | 3.40 | 0.0007 |
| x2 | 2.0122 | 0.7207 | 2.79 | 0.0052 |
Likelihood Ratio Statistics for y |
|||
| χ2 | d.f. | P | |
|---|---|---|---|
| x1 | 11.12 | 1 | 0.0009 |
| x2 | 7.85 | 1 | 0.0051 |
| TOTAL | 19.45 | 2 | <0.0001 |
| Imputation penalties | |||
| Test | Missing Information Fraction |
Denominator d.f. |
χ2 Discount |
|---|---|---|---|
| x1 | 0.218 | 10542.2 | 0.782 |
| x2 | 0.702 | 1011.3 | 0.298 |
| TOTAL | 0.541 | 3405.6 | 0.459 |
| Method: | Deletion | Single | Multiple |
|---|---|---|---|
| Allows non-random missing | x | x | |
| Reduces sample size | x | ||
| Apparent S.E. of \(\hat{\beta}\) too low | x | ||
| Increases real S.E. of \(\hat{\beta}\) | x | ||
| \(\hat{\beta}\) biased | if not MCAR | x |
The following contains crude guidelines. Simulation studies are needed to refine the recommendations. Here \(f\) refers to the proportion of observations having any variables missing.
NAs will be imputed first. aregImpute cycles in the order the analyst specifies variables in the formula.7 White et al. (2011) recommend choosing \(M\) so that the key inferential statistics are very reproducible should the imputation analysis be repeated. They suggest the use of \(100f\) imputations. See also (van Buuren, 2012, Section 2.7). von Hippel (2016) finds that the number of imputations should be quadratically increasing with the fraction of missing information.
Reason for missings more important than number of missing values.
Extreme amount of missing data does not prevent one from using multiple imputation, because alternatives are worse (Janssen et al., 2010; Madley-Dowd et al., 2019).
It is useful to look at examples of effective sample sizes in the presence of missing data. If a sample of 1000 subjects contains various amounts and patterns of missings what size \(n_c\) of a complete sample would have equivalent information for the intended purpose of the analysis?
Hmisc package aregImpute function and the rms stackMI function as demonstrated in the Titanic case study later in the notes.Section 3.1
Section 3.4
Section 3.5
Section 3.6
Section 3.8
Section 3.9
Section 3.10
Section 3.11
```{r include=FALSE}
options(qproject='rms', prType='html')
require(Hmisc)
require(qreport)
getRs('qbookfun.r')
hookaddcap()
knitr::set_alias(w = 'fig.width', h = 'fig.height', cap = 'fig.cap', scap ='fig.scap')
hooktime()
```
# Missing Data {#sec-missing-data}
`r quoteit("Consider a study of the effectiveness of a cardiac rehabilitation program. Suppose a sample size of N=300 will result in an acceptable margin of error in estimating the overall treatment effect. The investigators allow for 20% dropout so multiplies N by 1/0.8 = 1.25. What serious mistake are they likely making?")`
## Types of Missing Data
`r mrg(sound("missing-1"))`
* Missing completely at random (MCAR) `r ipacue()`
* Missing at random (MAR)^["Although missing at random (MAR) is a non-testable assumption, it has been pointed out in the literature that we can get very close to MAR if we include enough variables in the imputation models" @har07mul.]
* Informative missing (non-ignorable non-response; missing not at random; MNAR)
See @car21mis, @sch02mis, @don06rev, @har07mul, @all01mis, @whi11mul, @buu12fle for an
introduction to missing data and imputation concepts.
<!-- NEW: -->
See @haz23tri for a way to indirectly detect MNAR by comparing variances of response variables between treatments.
## Prelude to Modeling
* Quantify extent of missing data `r ipacue()`
* Characterize types of subjects with missing data
* Find sets of variables that are missing on same subjects
## Missing Values for Different Types of Response Variables
* Serial data with subjects dropping out (not covered in this `r ipacue()`
course^[@twi13mul found instability in using multiple imputation of longitudinal data, and advantages of using instead full likelihood models.]
<!-- TODO twi13mul is epub--->
* Excellent tutorial on imputation of longitudinal data: @wij25mul
* $Y$=time to event, follow-up curtailed: covered under survival
analysis^[@whi09imp provide a method for multiply imputing missing covariate values using censored survival time data.]
<!-- NEW following: -->
* Missing time-dependent covariates in longitudinal data: @mam23lon
* Often discard observations with completely missing $Y$ but sometimes wasteful^[$Y$ is so valuable that if one is only missing a $Y$ value, imputation is not worthwhile, and imputation of $Y$ is not advised if MCAR or MAR.]
* Characterize missings in $Y$ before dropping obs.
## Problems With Simple Alternatives to Imputation
Deletion of records---
* Badly biases parameter estimates when the probability of a `r ipacue()`
case being incomplete is related to $Y$ and not just
$X$ [@lit02sta].
* Deletion because of a subset of $X$ being missing
always results in inefficient estimates
* Deletion of records with missing $Y$ can result in
biases [@cra95com] but is the preferred approach
under MCAR^[Multiple imputation of $Y$ in that case does not improve the analysis and assumes the imputation model is correct.]
* @hip07reg found advantages to a "use
all variables to impute all variables then drop observations with
missing $Y$" approach (but see @sul15bia)
* @lee12rec suggest that observations missing
on both $Y$ and on a predictor of major interest are not helpful
* Only discard obs. when
+ MCAR can be justified
+ Rarely missing predictor of overriding importance that can't be
imputed from other data
+ Fraction of obs. with missings small and $n$ is large
* No advantage of deletion except savings of analyst time
* Making up missing data better than throwing away real data
* See @kno10unp
Adding extra categories of categorical predictors---
* Including missing data but adding a category `missing' causes `r ipacue()`
serious biases [@all01mis; @jon96ind; @vac98mis]
* Problem acute when values missing because subject too sick
* Difficult to interpret
* Fails even under MCAR [@jon96ind; @all01mis; @don06rev; @hei06imp; @kno10unp]
* May be OK if values are "missing" because of "not
applicable"^[E.g. you have a measure of marital happiness, dichotomized as high or low, but your sample contains some unmarried people. OK to have a 3-category variable with values high, low, and unmarried---Paul Allison, IMPUTE list, 4Jul09.]
* May be OK for pure prediction where interpretation of coefficients is of no interest and patterns of missingness will be the same in future data as in the training data
Likewise, serious problems are caused by setting missing continuous
predictors to a constant (e.g., zero) and adding an indicator variable
to try to estimate the effect of missing values.
Two examples from @don06rev using binary logistic
regression, $N=500$.
Results of 1000 Simulations With $\beta_{1}=1.0$ with MAR
and Two Types of Imputation `r ipacue()`
| Imputation | $\hat{\beta}_{1}$ | S.E. | Coverage of |
|-----|-----|-----|-----|
| Method | | | 0.90 C.I. |
| Single | 0.989 | 0.09 | 0.64 |
| Multiple | 0.989 | 0.14 | 0.90 |
Now consider a simulation with $\beta_{1}=1, \beta_{2}=0$, $X_{2}$
correlated with $X_{1} (r=0.75)$ but redundant in predicting $Y$, use
missingness indicator when $X_{1}$ is MCAR in 0.4 of 500 subjects.
This is also compared with grand mean fill-in imputation.
Results of 1000 Simulations Adding a Third Predictor Indicating Missing for $X_{1}$} `r ipacue()`
| Imputation | $\hat{\beta}_{1}$ | $\hat{\beta}_{2}$ |
|-----|-----|-----|
| Method | | |
| Indicator | 0.55 | 0.51 |
| Overall mean| 0.55 | |
In the incomplete observations the constant $X_{1}$ is uncorrelated
with $X_{2}$.
## Strategies for Developing an Imputation Model
**The goal of imputation is to preserve the information and meaning of the non-missing data.**
`r mrg(sound("missing-2"))`
There is a full Bayesian modeling alternative to all the methods
presented below. The Bayesian approach requires more effort but has
several advantages [@erl16dea].
Exactly how are missing values estimated?
* Predictive model for each target uses any outcomes, all
predictors in the final model other than the target, plus
auxiliary variables not in the outcome model
* Could ignore all other information --- random or grand mean `r ipacue()`
fill-in
* Can use external info not used in response model (e.g., zip
code for income)
* Need to utilize reason for non-response if possible
* Use statistical model with sometimes-missing $X$ as response
variable
* Model to estimate the missing values should include all
variables that are either `r ipacue()`
1. related to the missing data mechanism;
1. have distributions that differ between subjects that have the
target variable missing and those that have it measured;
1. associated with the sometimes-missing variable when it is not
missing; or
1. included in the final response model [@bar04imp; @har07mul]
* Ignoring imputation results in biased $\hat{V}(\hat{\beta})$
* `transcan` function in Hmisc library: "optimal"
transformations of all variables to make residuals more stable and
to allow non-monotonic transformations
* `aregImpute` function in Hmisc: good approximation to full
Bayesian multiple imputation procedure using the bootstrap
* `transcan` and `aregImpute` use the following for fitting
imputation models: `r ipacue()`
1. initialize `NA`s to median (mode for categoricals)
1. expand all categorical predictors using dummy variables
1. expand all continuous predictors using restricted cubic splines
1. optionally optimally transform the variable being predicted by
expanding it with restricted cubic splines and using the first
canonical variate (multivariate regression) as the optimum
transformation (maximizing $R^2$)
1. one-dimensional scoring of categorical variables being
predicted using canonical variates on dummy variables representing
the categories (Fisher's optimum scoring algorithm); when imputing
categories, solve for which category yields a score that is
closest to the predicted score
* `aregImpute` and `transcan` work with `r ipacue()`
`fit.mult.impute` to make final analysis of response variable
relatively easy
* Predictive mean matching
* Recursive partitioning with surrogate splits --- handles case
where a predictor of a variable needing imputation is missing
itself. But there are problems [@pen18pro] even with completely
random missingness.
* @whi11mul discusses an alternative method based on
choosing a donor observation at random from the $q$ closest matches
($q=3$, for example)
### Interactions
* When interactions are in the outcome model, oddly enough it may `r ipacue()`
be better to treat interaction terms as "just another variable"
and do unconstrained imputation of them [@kim15eva]
## Single Conditional Mean Imputation
`r mrg(sound("missing-3"))`
* Can fill-in using unconditional mean or median if number of `r ipacue()`
missings low and $X$ is unrelated to other $X$s
* Otherwise, first approximation to good imputation uses other
$X$s to predict a missing $X$
* This is a single "best guess" conditional mean
* $\hat{X}_{j} = Z \hat{\theta}, Z = X_{\bar j}$ plus possibly
auxiliary variables that precede $X_{j}$ in the causal chain that
are not intended to be in the outcome model.<br>
Cannot include $Y$ in $Z$ without adding random errors to imputed
values as done with multiple imputation (would steal info from $Y$)
* Recursive partitioning can sometimes be helpful for nonparametrically
estimating conditional means
## Predictive Mean Matching
* Developed by @lit02sta
* Replace missing value with observed value of subject having closest predicted
value to the predicted value of the subject with the `NA`.
Key considerations are how to
1. model the target when it is not `NA`
1. match donors on predicted values
1. avoid overuse of "good" donors to disallow excessive ties in
imputed data
1. account for all uncertainties
* No distributional assumptions; nicely handles target variables
with strange distributions [@vin14pre]
* Predicted values need only be monotonically related to real
predictive values
+ PMM can result in some donor observations being used repeatedly `r ipacue()`
+ Causes lumpy distribution of imputed values
+ Address by sampling from multinomial distribution,
probabilities = scaled distance of all predicted values to
predicted value ($y^{*}$) of observation needing imputing
+ Tukey's tricube function is a good weighting function (used in
loess): $w_{i} = (1 - \min(d_{i}/s, 1)^{3})^{3}$, <br>
$d_{i} = |\hat{y_{i}} - y^{*}|$ <br>
$s = 0.2\times\text{mean} |\hat{y_{i}} - y^{*}|$ is a good default scale factor. <br>
Scale so that $\sum w_{i} = 1$
<!-- NEW -->
### Predictive Mean Matching With Constraints {#sec-missing-constraints}
* PMM is empirical so it is relatively easy to add constraints
+ Need sample size to be large enough so that there are several donor observations meeting the constraints
* Implemented in `Hmisc` 5.1-1
* For each variable to be imputed the user can specify an R expression that is a logical TRUE/FALSE condition specifying which observations qualify
* Constraint can involve relationships between potential donor observations (variables prefixed by `d$`) and the recipient (target) observation (variables prefixed by `r$`) having the missing value
* PMM on the qualifying donor observations
* Example
+ time 0 corresponds to the day a patient is admitted to the hospital
+ day of discharge is missing for some patients
+ every patient had at least one follow-up visit
+ constrain the day of discharge to be before the day of the first follow-up visit
```{r eval=FALSE}
a <- aregImpute(~ age + day_dis + day_follow_up, data=d,
constraint=list(day_dis=expression(d$day_dis < r$day_follow_up)))
```
## Multiple Imputation
* Single imputation could use a random draw from the conditional `r ipacue()`
distribution for an individual <br>
$\hat{X}_{j} = Z \hat{\theta} + \hat{\epsilon}, Z = [X\bar{j},
Y]$ plus auxiliary variables <br>
$\hat{\epsilon} = n(0, \hat{\sigma})$ or a random draw from the
calculated residuals
+ bootstrap
+ approximate Bayesian bootstrap [@rub91mul; @har07mul]: sample
with replacement from sample with replacement of residuals
* Multiple imputations ($M$) with random draws
+ Draw sample of $M$ residuals for each missing value to be imputed
+ Average $M$ $\hat{\beta}$
+ In general can provide least biased estimates of $\beta$
+ Simple formula for imputation-corrected
var($\hat{\beta}$) <br> Function of average "apparent"
variances and between-imputation variances of
$\hat{\beta}$
+ Even when the $\chi^2$ distribution is a good approximation
when data have no missing values, the $t$ or $F$ distributions are
needed to have accurate $P$-values and confidence limits when
there are missings [@lip02deg; @rei07sma]
+ **BUT** full multiple imputation needs to account for
uncertainty in the imputation models by refitting these models for
each of the $M$ draws
+ `transcan` does not do that; `aregImpute` does
* Note that multiple imputation can and should use the response
variable for imputing predictors [@moo06usi]
* `aregImpute` algorithm [@moo06usi] `r ipacue()`
`r mrg(sound("missing-4"))`
+ Takes all aspects of uncertainty into account using the
bootstrap
+ Different bootstrap resamples used for each imputation by fitting
a flexible additive model on a sample with replacement
from the original data
+ This model is used to predict all of the original missing and
non-missing values for the target variable for the current imputation
+ Uses flexible parametric additive regression models to impute
+ There is an option to allow target variables to be optimally
transformed, even non-monotonically (but this can overfit)
+ By default uses predictive mean matching for imputation; no residuals
required (can also do more parametric regression imputation)
+ By default uses weighted PMM; many other matching options
+ Uses by default van~Buuren's "Type 1" matching \cite[Section
3.4.2]{buu12fle} to capture the right amount of uncertainty by
computing predicted values for missing values using a
regression fit on the bootstrap sample, and finding donor
observations by matching those predictions to predictions from potential
donors using the regression fit from the original sample
of complete observations
+ Uses Rubin's rule to estimate variances of $\hat{\beta}$, involving between- and within-imputation variance combination
+ When a predictor of the target variable is missing, it is first
imputed from its last imputation when it was a target variable
+ First 3 iterations of process are ignored ("burn-in")
+ Compares favorably to `R` `MICE` approach
+ Example:
```{r eval=FALSE,timeit=FALSE}
a <- aregImpute(~ age + sex + bp + death + heart.attack.before.death,
data=mydata, n.impute=5)
f <- fit.mult.impute(death ~ rcs(age,3) + sex +
rcs(bp,5), lrm, a, data=mydata)
```
See @bar04imp for a nice review of multiple
imputation with detailed comparison of results
(point estimates and confidence limits for the effect of the
sometimes-missing predictor) for various imputation methods.
@bar06mul have a good overview of imputation
methods and a comparison of bias and confidence interval coverage for
the methods when applied to longitudinal data with a small number of subjects.
@hor07muc have a good review of several
software packages for dealing with missing data, and a comparison of
them with `aregImpute`. @har07mul provide a
nice overview of multiple imputation and discuss some of the
available software. @whi10bia studied bias
of multiple imputation vs. complete-case analysis.
@whi11mul provide much practical guidance.
**Caution**: Methods can generate imputations having very
reasonable distributions but still not having the property that final
response model regression coefficients have nominal confidence
interval coverage. It is worth checking that imputations generate the
correct collinearities among covariates.
* With `MICE` and `aregImpute` we are using the chained `r ipacue()`
equation approach [@whi11mul] `r ipacue()`
* Chained equations handles a wide variety of target variables to
be imputed and allows for multiple variables to be missing on the
same subject
* Iterative process cycles through all target
variables to impute all missing values [@buu06ful]
* Does not attempt to use the full Bayesian
multivariate model for all target variables, making it more
flexible and easy to use
* Possible to create improper imputations, e.g., imputing
conflicting values for different target variables
* However, simulation studies [@buu06ful] demonstrate
very good performance of imputation based on chained equations
<!-- NEW -->
## Likelihood Ratio Tests and Multiple Imputation {#sec-missing-lrt}
* For frequentist-based analysis Wald tests are ultimately unsatisfying for reasons detailed in @sec-mle-hd
+ Also it was never clear that simple averaging of $\hat{\beta}$ over completed datasets is the best approach
* @cha22mul have developed a promising approach to obtaining approximate LRT (likelihood ratio tests) in the presence of missing data
+ [Web site](https://sites.google.com/site/kwchankeith/publications/milrt)
+ [arXiv paper](https://arxiv.org/abs/1711.08822)
+ [R code](https://drive.google.com/file/d/1mHVT3yV2oX1TCHR9zZgC6XIIwAaB3Gtv/view?usp=sharing)
+ See @woo08how for related work
* Instead of averaging $M$ $\hat{\beta}$ from multiple separate analyses of completed datasets, stacks all completed datasets into one large dataset and computes maximum likelihood estimates on the one large stacked dataset
* Divide LR $\chi^2$ by $M$ to get a rough estimate of the more correct LR described below; need to multiply by a discounting factor to take imputation into account
* Individual fit LRs are used to derive imputation correction factors and the fraction of missing information
* Chan and Meng have a formula that provides more accuate $p$-values by solving for denominator degrees of freedom for an $F$ distribution
+ The degrees of freedom must be computed separately for each hypothesis test
+ Accounts for heavier tail problem mentioned earlier
+ $F$ test denominator d.f. is $\frac{k (M - 1)}{\hat{f}^{2}}$ where $k$ is the numerator d.f. (number of parameters being tested simultaneously), the fraction of missing information $\hat{f} = \frac{\hat{r}}{1+\hat{r}}$, $\hat{r} = \max(0, \frac{M+1}{k(M-1)} (\bar{d} - \hat{d}))$, where $\bar{d}$ is the mean over imputed datasets of the individual LR statistics and $\hat{d}$ is LR on the stacked dataset, divided by $M$
+ Use large $M$ to make the $\chi^2$ approximation better by making the denominator d.f. larger. E.g., if $k=1$ and $\hat{f}=0.5$, $M=26$ already yields more than 100 denominator d.f. for $F$, making it hard to distinguish from a $\chi_{1}^{2}$ distribution. If the test statistic is 4.0, the $p$-value $\chi_{1}^{2}$ is 0.0455 and is 0.048 from $F_{1, 100}$.
* `Hmisc` `fit.mult.impute` function run with `lrt=TRUE` runs and saves all the needed LR tests to allow `rms` `processMI` to compute everything else
* `processMI` computes $\hat{f}$, the $F$ test denominator d.f., and the $\chi^2$ discount factor and uses these to get the final imputation-adjusted LR tests
* `fit.mult.impute` computes very approximate Wald statistics by multiplying the variance-covariance matrix computed on the stacked data by $M$ if individual completed dataset analyses are not done
* Chan and Meng approach uses $\hat{\beta}$ computed once on stacked data as the MLE in the multiple imputation context; `fit.mult.impute` computes the final $\hat{\beta}$ this way if stacking is done; however the variance-covariance matrix is estimated from Rubin's rule as in the Wald approach first used in this chapter
* Imputation-adjusted LRTs can be a large computational burden for large datasets---for $M$ imputations, $j$ effects being tested requires $(M+1) j$ model fits. For each imputation, the `rms` `anova` function has very litle overhead though, due to it only computing the design matrix once, and calling low-level fitting functions for all LRTs once the overall model is fitted with a high-level function.
**Example**
```{r}
require(rms)
set.seed(1)
n <- 500
x1 <- runif(n)
x2 <- runif(n)
L <- x1 + 2*x2
y <- rbinom(n, 1, plogis(L))
x2[1:300] <- NA # make 300 observations have missing x2
d <- data.frame(x1, x2, y)
a <- aregImpute(~ y + x1 + x2, n.impute=10, data=d, pr=FALSE)
f <- fit.mult.impute(y ~ x1 + x2, lrm, a, data=d, pr=FALSE)
f
```
In the above results, the LR $\chi^2$ did not account for imputation and is too high. So are the $R^2$ measures derived from it.
Compute Wald $\chi^2$ test statistics accounting for imputation
```{r}
anova(f)
```
Now compute LR test statistics accounting for imputation
```{r timeit=TRUE}
# lrt=TRUE makes fit.mult.impute set up all the arguments needed for LR tests
h <- fit.mult.impute(y ~ x1 + x2, lrm, a, data=d, lrt=TRUE, pr=FALSE)
h
as <- processMI(h, which='anova') # LR tests accounting for imputation
as
prmiInfo(as)
```
The Chan & Meng $F$ approximation (`processMI` uses the $\chi^2$ version of it) is needed for `x2` as the denominator d.f. of 14.5 is too low for $p$-values from the $\chi^2$ distribution to well approximate those from the $F$ distribution. Increase the number of imputations to increase the denominator d.f.
```{r timeit=TRUE}
a <- aregImpute(~ y + x1 + x2, n.impute=50, data=d, pr=FALSE)
h <- fit.mult.impute(y ~ x1 + x2, lrm, a, data=d, lrt=TRUE, pr=FALSE)
h
as <- processMI(h, which='anova')
as
prmiInfo(as)
```
What happens if we use an extremely high number of imputations?
```{r timeit=TRUE}
a <- aregImpute(~ y + x1 + x2, n.impute=500, data=d, pr=FALSE)
h <- fit.mult.impute(y ~ x1 + x2, lrm, a, data=d, lrt=TRUE, pr=FALSE)
h
as <- processMI(h, which='anova')
as
prmiInfo(as)
```
## Diagnostics
`r mrg(sound("missing-5"))`
* MCAR can be partially assessed by comparing distribution of `r ipacue()`
non-missing $Y$ for those subjects with complete $X$ vs. those
subjects having incomplete $X$ [@lit02sta]
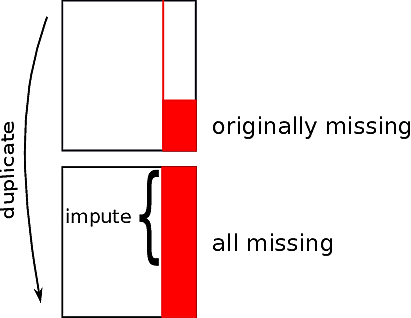
* Yucel and Zaslavsky [@yuc08usi; see also @he12dia]
* Interested in reasonableness of imputed values for a
sometimes-missing predictor $X_{j}$
* Duplicate entire dataset
* In the duplicated observations set all non-missing values of
$X_{j}$ to missing; let $w$ denote this set of observations set to missing
* Develop imputed values for the missing values of $X_{j}$
* In the observations in $w$ compare the
distribution of imputed $X_{j}$ to the original values of $X_{j}$
* @bon16gra present a variety of
useful diagnostics on the reasonableness of imputed values.
<img src="images/missing-diagnostic.png" width="60%">
## Summary and Rough Guidelines
`r mrg(sound("missing-6"))` `r ipacue()`
| Method: | Deletion | Single | Multiple |
|-----------------------------------------|---------------|---|---|
| Allows non-random missing | | x | x |
| Reduces sample size | x | | |
| Apparent S.E. of $\hat{\beta}$ too low | | x | |
| Increases real S.E. of $\hat{\beta}$ | x | | |
| $\hat{\beta}$ biased | if not MCAR | x | |
: Summary of methods for dealing with missing values {#tbl-na-meth-summary}
The following contains crude guidelines. Simulation studies are
needed to refine the recommendations. Here $f$ refers to
the proportion of observations having _any_ variables missing.
* **$f < 0.03$:** `r ipacue()`
It doesn't matter very much how you impute missings or whether
you adjust variance of regression coefficient estimates for
having imputed data in this case. For
continuous variables imputing missings with the median
non-missing value is adequate; for categorical predictors the
most frequent category can be used. Complete case analysis is
also an option here. Multiple imputation may be needed to check
that the simple approach "worked."
* **$f \geq 0.03$:**
Use multiple imputation with number of
imputations^[@whi11mul recommend choosing $M$ so that the key inferential statistics are very reproducible should the imputation analysis be repeated. They suggest the use of $100f$ imputations. See also [@buu12fle, section 2.7]. @hip16num finds that the number of imputations should be quadratically increasing with the fraction of missing information.]
equal to $\max(5, 100f)$. Fewer imputations may be possible with
very large sample
sizes. See [statisticalhorizons.com/how-many-imputations](https://statisticalhorizons.com/how-many-imputations).
Type 1 predictive mean matching is usually preferred, with
weighted selection of donors. Account for imputation in estimating
the covariance matrix for final parameter estimates. Use the $t$
distribution instead of the Gaussian distribution for tests and
confidence intervals, if possible, using the estimated d.f. for the
parameter estimates.
* **Multiple predictors frequently missing:** `r ipacue()`
More imputations may be required. Perform a "sensitivity to
order" analysis by creating multiple imputations
using different orderings of sometimes missing variables. It may be
beneficial to initially sort variables so that the one with the most
`NA`s will be imputed first. `aregImpute` cycles in the order the analyst specifies variables in the formula.
Reason for missings more important than number of missing values.
Extreme amount of missing data does not prevent one from using
multiple imputation, because alternatives are worse [@jan10mis; @mad19pro].
### Effective Sample Size
It is useful to look at examples of effective sample sizes in the presence of missing data. If a sample of 1000 subjects contains various amounts and patterns of missings what size $n_c$ of a complete sample would have equivalent information for the intended purpose of the analysis?
1. A new marker was collected on a random sample of 200 of the subjects and one wants to estimate the added predictive value due to the marker: $n_{c}=200$ `r ipacue()`
1. Height is missing on 100 subjects but we want to study association between BMI and outcome. Weight, sex, and waist circumference are available on all subjects: $n_{c}=980$
1. Each of 10 predictors is randomly missing on $\frac{1}{10}$ of subjects, and the predictors are uncorrelated with each other and are each weakly related to the outcome: $n_{c}=500$
1. Same as previous but the predictors can somewhat be predicted from non-missing predictors: $n_{c}=750$
1. The outcome variable was not assessed on a random $\frac{1}{5}$ of subjects: $n_{c}=800$
1. The outcome represents sensitive information, is missing on $\frac{1}{2}$ of subjects, and we don't know what made subjects respond to the question: $n_{c}=0$ (serious selection bias)
1. One of the baseline variables was collected prospectively $\frac{1}{2}$ of the time and for the other subjects it was retrospectively estimated only for subjects ultimately suffering a stroke and we don't know which subjects had a stroke: $n_{c}=0$ (study not worth doing)
1. The outcome variable was assessed by emailing the 1000 subjects, for which 800 responded, and we don't know what made subjects respond: $n_{c}=0$ (model will possibly be very biased---at least the intercept)
<!-- NEW -->
## Bayesian Methods for Missing Data
* Multiple imputation developed as an approximation to a full `r ipacue()`
Bayesian model
* Full Bayesian model treats missings as unknown parameters and
provides exact inference and correct measures of uncertainty
* See [this case study](https://github.com/paul-buerkner/brms/blob/master/vignettes/brms_missings.Rmd) for an example
* The case study also shows how to do "posterior stacking" if
you want to avoid having to specify a full model for missings, and
instead use usual multiple imputations as described in this chapter. See @zho12not.
+ Run a multiple imputation algorithm
+ For each completed dataset run the Bayesian analysis and draw
thousands of samples from the posterior distribution of the parameters
+ Pool all these posterior draws over all the multiple
imputations and do posterior inference as usual with no special
correction required
+ Made easy by the `Hmisc` package `aregImpute` function
and the `rms` `stackMI` function as demonstrated in the
Titanic case study later in the notes.
## Study Questions
**Section 3.1**
1. What is the problem with doing ordinary analysis of data from survey responders?
**Section 3.4**
1. What problem is always present when doing complete-case analysis when missing values exist in the data?
1. What problem is often present?
1. Why does imputation not help very much when a variable being imputed is a main variable of interest in the analysis?
1. What is a major reason that adding a new category for a predictor for missings doesn't work?
1. Why does inserting a constant for missing values of a continuous predictor primarily fail?
**Section 3.5**
1. What is a more accurate statement than "imputation boosts the sample size"?
1. What does single-value fill-in of missings almost always damage?
1. What is a general way to describe why predictive mean matching (PMM) works?
1. What is a general advantage of PMM?
**Section 3.6**
1. Why does single conditional mean imputation result in biased regression coefficients?
**Section 3.8**
1. Why can multiple imputation use Y in predicting X?
1. What are the sources of uncertainty that a multiple imputation algorithm must take into account for final standard errors to not be underestimated?
**Section 3.9**
1. Explain the Yucel-Zaslavsky diagnosic and what it is checking for.
**Section 3.10**
1. What is the only reason not to always do 100 or more imputations?
**Section 3.11**
1. If using multiple imputation but within a Bayesian framework, what is a major advantage of posterior stacking over what we have been doing in the frequentist domain?